Translational Epigenetics Laboratory
AG Papantonis
Our research focuses on dissecting the structure-to-function relationship of the human genome at the molecular level. We wish to understand how chromatin integrates the various signaling stimuli of its environment to control transitions between homeostatic and deregulated functional programs. We particularly ask how changes along the linear DNA fiber translate into dynamic higher-order regulatory networks. Ultimately, by deciphering the general rules governing transcriptional and chromatin homeostasis, we will be able to compile a parsimonious set of rules that allows prediction of how a cell might respond during development, in ageing or upon malignancy.
What we do (for the non-scientific audience)
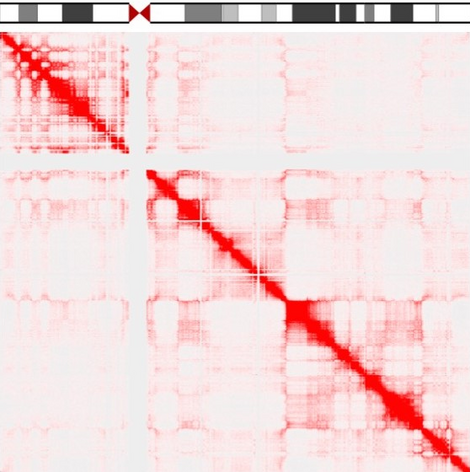
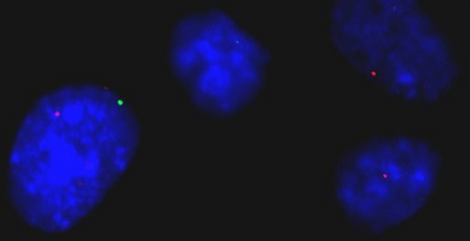
The efforts of the Human Genome Project have revealed the sequence of the billions of bases in our chromosomes. However, how these very long molecules fold in 3D space, and how this folding allows our genes to be expressed at the right time and place, still eludes us. We are currently trying to generate 3D maps of the human genome to identify gene neighborhoods. This should allow us to understand the rules that govern gene function, and to predict how a cell might respond in disease or ageing.
Selected publications
- Frank S, Ahuja G, Bartsch D, Russ N, Yao W, Kuo JC, Derks JP, Akhade VS, Kargapolova Y, Georgomanolis T, Messling JE, Gramm M, Brant L, Rehimi R, Vargas NE, Kuroczik A, Yang TP, Sahito RGA, Franzen J, Hescheler J, Sachinidis A, Peifer M, Rada-Iglesias A, Kanduri M, Costa IG, Kanduri C, Papantonis A, Kurian L (2019) yylncT defines a class of divergently transcribed lncRNAs and safeguards the T-mediated mesodermal commitment of human PSCs. Cell Stem Cell 24: 318-327
- Rada-Iglesias A*, Grosveld FG, Papantonis A*. Forces driving the three-dimensional folding of eukaryotic genomes. Mol Syst Biol 14: e8214 (*co-corresponding authors)
- Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmüller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nürnberg P, Marenduzzo D, Rippe K, Papantonis A (2018) HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell 70: 730-744
- Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmüller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nürnberg P, Marenduzzo D, Rippe K, Papantonis A (2018) HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell 70: 730-744
- Michieletto D, Chiang M, Colì D, Papantonis A, Orlandini E, Cook PR, Marenduzzo D (2018) Shaping epigenetic memory via genomic bookmarking. Nucleic Acids Res 46: 83-93
- Franzen J, Zirkel A, Blake J, Rath B, Benes V, Papantonis A, Wagner W (2017) Senescence-associated DNA methylation is stochastically acquired in subpopulations of mesenchymal stem cells. Aging Cell 16: 183-91
- Brant L, Georgomanolis T, Nikolic M, Brackley CA, Kolovos P, van Ijcken W, Grosveld FG, Marenduzzo D, Papantonis A* (2016) Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Mol Syst Biol 12: 891 (*corresponding author)
- Nikolic M, Papantonis A, Rada-Iglesias A (2016) GARLiC: A bioinformatic toolkit for etiologically connecting diseases and cell type-specific regulatory maps. Hum Mol Genet doi: 10.1093/hmg/ddw423
- Kolovos P*, Georgomanolis T, Koeferle A, Larkin JD, Brant L, Nikolic M, Gusmao EG, Zirkel A, Knoch TA, van Ijcken W, Cook PR, Costa IG, Grosveld FG, Papantonis A* (2016) Binding of nuclear factor kappaB to noncanonical consensus sites reveals its bimodal role during the early proinflammatory response. Genome Res 26: 1478-89 (*corresponding author)
- Melnik S, Caudron-Herger M, Brant L, Rippe K, Cook PR, Papantonis A* (2015) Isolation of the protein and RNA content of the active site of transcription in mammalian cells. Nat Protocols 11: 553-65 (*corresponding author)
- Caudron-Herger M, Cook PR, Rippe K, Papantonis A* (2015) Dissecting the nascent human transcriptome by analyzing the RNA content of transcription factories. Nucleic Acids Res 43: e95 (*corresponding author)
- Kelly S, Georgomanolis T, Zirkel A, Diermeier S, O’Reilly D, Murphy S, Laengst G, Cook PR, Papantonis A* (2015) Splicing of many human genes involves sites embedded within introns. Nucleic Acids Res 43: 4721-32 (*corresponding author)
- Kelly S, Greenman C, Cook PR, Papantonis A* (2015) Exon skipping is correlated with exon circularization. J Mol Biol 427: 2414-7 (*corresponding author)
- Diermeier S, Kolovos P, Heizinger L, Schwartz U, Georgomanolis T, Zirkel A, Wedemann G, Grosveld F, Knock TA, Merckl R, Cook PR, Laengst G*, Papantonis A* (2014) TNFα signaling primes chromatin for NF-κB binding and induces rapid and widespread nucleosome repositioning. Genome Biol 15: 536 (*co-corresponding authors)
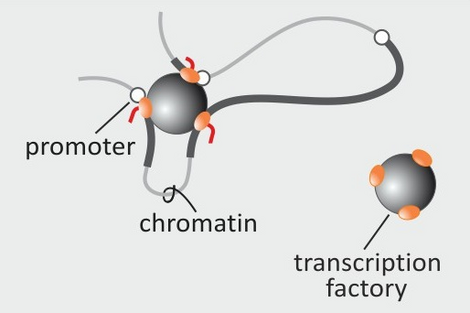
- Papantonis A, Cook PR (2013) Transcription factories: genome organization and gene regulation. Chem Rev doi:10.1021/ cr300513p
- Larkin JD, Cook PR, Papantonis A* (2012) Dynamic configurations of long human genes during a transcription cycle. Mol Cell Biol 32: 2738-2747 (*corresponding author)
For a complete list of publications by our group, please visit: https://www.ncbi.nlm.nih.gov/pubmed/?term=papantonis+a
Lab members

Argyris Papantonis (b. 1978, Athens, Greece) studied Biology at the National and Kapodistrian University of Athens, where he also undertook his PhD studies under the supervision of Prof. Rena Lecanidou. In 2008 he moved to Oxford for his post-doctoral work with Prof. Peter Cook at the Sir William Dunn School of Pathology. In 2013 he was appointed as a Junior Research Group Leader for Chromatin Systems Biology at the CMMC, and in 2018 he was recruited to the Medical Faculty of the Georg-August University of Göttingen where he is currently a W2-Professor for Translational Epigenetics. When he is not doing research he reads, writes, or translates literary texts.
E-mail: argyris.papantonis [at] med.uni-goettingen.de / Twitter: @apapant
Current lab members:
Dr. Athanasia Mizi (Senior postdoc) studied Biology at the National and Kapodistrian University of Athens, where she also undertook her PhD studies under the supervision of Prof. George Rodakis on the molecular evolution of molluscan mtDNA. Before joining our lab, Athanasia did her first post-doctoral work with Dr. Paul Brinkkoetter on the role of the key mitochondrial factor, TFAM, in kidney function. Currently Athanasia is working on miRNA-centered 3D networks; she is funded by the UMG.
E-mail: athanasia.mizi [at] med.uni-goettingen.de
Dr. Eduardo Gade Gusmao (Bioinformatician) initially studied Computer Sciences in Pernambuco, Brazil, before undertaking his PhD work with Ivan Costa at the RWTH University of Aachen, Germany, and developing an interest in transcription factor binding properties on chromatin. Eduardo joined our laboratory in February 2018 and is interested in deep learning and in new tools that further exploit native Hi-C data; he is funded via the SPP 2202 Priority Program.
E-mail: eduardo.gusmao [at] med.uni-goettingen.de
Dr. Ting Xie (postdoc) studied Bioinformatics in the Huazhong Agricultural University, China, where she also undertook her MSc studies. Before joining our lab, Ting worked as an Assistant Professor in Oil Crops Research Institute, Chinese Academy of Agricultural Sciences studying on Brassica 3D genome and evolution. Currently, Ting is working on structural variation in tumor. She is funded by Humboldt Fellowship by Alexander von Humboldt Foundation.
E-mail: ting.xie(at)med.uni-goettingen.de
Dr. Mariano Barbieri (Physicist) graduated in Theoretical Physics in "Federico II" University of Napoli, Italy, where he also dedicated the PhD to Statistical Mechanics models of chromatin architecture with Prof. Mario Nicodemi. Later he spent the postdoc time in Prof. Ana Pombo's lab in Berlin. He joined our laboratory in 2021 after an experience in the private sector (quantitative finance at Morgan Stanley; optics simulations at Italian Space Agency) to investigate models of nuclear organization. He's interested in complex systems behavior and machine learning techniques.
E-mail: mariano.barbieri(at)med.uni-goettingen.de
Natasa Josipovic (PhD student) studied Biology at the University of Belgrade, where she also undertook her MSc studies. Her placement as a PhD student started in 2016, and she has been working on single-nulceotide resolution and single-cell nascent RNA-seq to understand fundamental differences in the temporal deployment of TNF- versus TGF-driven transcriptional responses; she is funded by a DFG grant.
E-mail: natasa.josipovic [at] med.uni-goettingen.de
Shu Zhang (PhD student) studied Agricultural Sciences in China, where she also did an MSc specialized in bioinformatics. Her placement as a PhD student started in late 2018, and she is working on genome reogranziation upon aging; she is supported by a full CAAS scholarship.
E-mail: shu.zhang [at] med.uni-goettingen.de
Nadine Übelmesser (PhD student) studied Biology in Erlangen, where she also undertook her MSc studies on Cellular and Molecular Biology. Her placement as a PhD student started in 2019, and she is working on cell cycle progression and its link to chromosomal folding in cancer; she is funded via TRR81/SFB program.
E-mail: nadine.uebelmesser [at] med.uni-goettingen.de
Spyridon Palikyras (PhD student) studied Biology in Greece and received his MSc degree at the University of Heidelberg. His placement as a PhD student started in 2019, and he is working on phase-separated CTCF clustering in cellular senescence; he is funded via TRR81/SFB program.
E-mail: spyridon.palikyras [at] med.uni-goettingen.de
Adi Mackay (PhD student) studied Applied Biology in HBRS, and MSc Molecular Medicine in the UMG. She did her MSc thesis in our lab working on probing genome reorganization via ligation-/crosslinking-free methods in live cells and currently pursuing her PhD by investigating pancreatic cancer through the patient derived organoids model. She is funded by the KFO5002 program.
E-Mail: adi.mackay(at)med.uni-goettingen.de
Yajie Zhu (PhD student) has been in IMPRS MSc/PhD program Molecular Biology, Goettingen, since 2018. After completing her Master thesis in our lab, she started her PhD study in Dec., 2020, co-supervised by Prof. Johannes Soeding. She is working on genome reorganization during tumorigenesis by analyzing high throughput sequencing data.
E-mail: yajie.zhu@med.uni-goettingen.de
Alumni:
- Dr. Anne Zirkel (former postdoc), currently holding a managerial position in the private healthcare sector.
- Dr. Lilija Brant (former PhD student), currently holding a consultant in the private sector.
- Dr. Milos Nikolic (former PhD student), currently a postdoc with the Peifer lab in Cologne
- Dr. Konstantinos Sofiadis (former PhD student), currently a postdoc with the de Laat lab in Utrecht.
Contact
How to find us? We are currently located on the 2nd floor of the main UMG building (Room 2.E1.216).
Postal address:
AG Papantonis: Translational Epigenetics
Institute of Pathology, University Medical Center,
Georg-August University of Göttingen,
Robert-Koch-Str. 40, 37075 Göttingen, Germany
Tel.: +49 551 39 65734

Kontaktinformationen
- Telefon: +49 551 3965734
- E-Mail-Adresse: argyris.papantonis(at)med.uni-goettingen.de
Das könnte Sie auch interessieren
