Translational Epigenetics Laboratory
AG Papantonis
Our research focuses on dissecting the structure-to-function relationship of the human genome at the molecular level. We wish to understand how chromatin integrates the various signaling stimuli of its environment to control transitions between homeostatic and deregulated functional programs. We particularly ask how changes along the linear DNA fiber translate into dynamic higher-order regulatory networks. Ultimately, by deciphering the general rules governing transcriptional and chromatin homeostasis, we will be able to compile a parsimonious set of rules that allows prediction of how a cell might respond during development, in ageing or upon malignancy.
What we do (for the non-scientific audience)
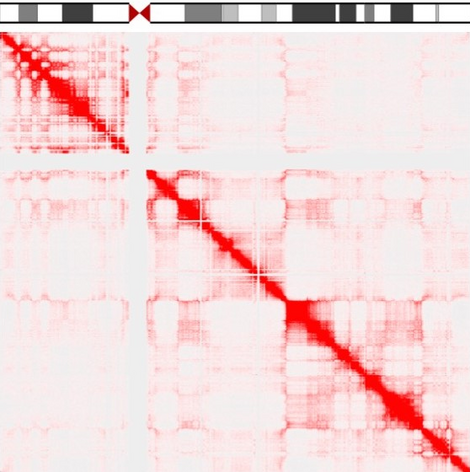
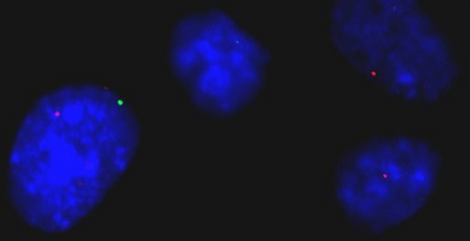
The efforts of the Human Genome Project have revealed the sequence of the billions of bases in our chromosomes. However, how these very long molecules fold in 3D space, and how this folding allows our genes to be expressed at the right time and place, still eludes us. We are currently trying to generate 3D maps of the human genome to identify gene neighborhoods. This should allow us to understand the rules that govern gene function, and to predict how a cell might respond in disease or ageing.
Selected publications
- Longo GMC, Sayols S, Stefanova ME, Xie T, Elsayed W, Panagi A, Stavridou AI, Petrosino G, Ing-Simmons E, Melo US, Gothe HJ, Vaquerizas JM, Kotini AG, Papantonis A*, Mundlos S*, Roukos V*. Type II topoisomerases shape multi-scale 3D chromatin folding in regions of positive supercoils. Mol Cell doi:10.1016/j.molcel.2024.10.007.
- Xie T, Danieli-Mackay A, Buccarelli M, Barbieri M, Papadionysiou I, D’Alessandris QG, Übelmesser N, Vinchure OS, Lauretti L, Fotia G, Wang X, Ricci-Vitiani L, Gopalakrishnan J, Pallini R, Papantonis A* (2024) Extreme structural heterogeneity rewires glioblastoma chromosomes to sustain patient-specific transcriptional programs. Nat Commun doi:10.1038/s41467-024-48053-2.
- Zhang S, Übelmesser N, Barbieri M, Papantonis A* (2023) Enhancer-promoter contact formation requires RNAPII and antagonizes loop extrusion. Nat Genet doi:10.1038/s41588-023-01364-4. [Highlighted in Nat Genetics]
- Debès C, Papadakis A, Karalay Ö, Tain L, Mizi A, Nakamura S, Hahn O, Weigelt C, Josipovic N, Zirkel A, Brusius I, Sofiadis K, Lamprousi M, Lu YX, Huang W, Esmaillie R, Kubacki T, Späth MR, Schermer B, Benzing T, Müller RU, Antebi A*, Partridge L*, Papantonis A*, Beyer A* (2023) Aging-associated changes in transcriptional elongation influence metazoan longevity. Nature doi:10.1038/s41586-023-05922-y. [Highlighted by AAAS EurekaAlert!]
- Zampetidis CP, Galanos P, Angelopoulou A, Zhu Y, Karamitros T, Polyzou A, Karamitros T, Kotsinas A, Lagopati N, Mourkioti I, Mirzazadeh R, Polyzos A, Garnerone S, Mizi A, Gusmao EG, Sofiadis K, Gal Z, Larsen DH, Pefani DE, Demaria M, Tsirigos A, Crosetto N, Maya-Mendoza A, Papaspyropoulos A, Evangelou K, Bartek J*, Papantonis A*, Gorgoulis VG* (2021) A recurrent chromosomal inversion suffices for driving escape from oncogene-induced senescence via subTAD reorganization. Mol Cell 81:4907-4923. [Highlighted by Cancer Research]
- Zhang S, Übelmesser N, Josipovic N, Forte G, Slotman JA, Chiang M, Gothe H, Gusmao EG, Becker C, Altmüller J, Houtsmuller AB, Roukos V, Wendt KS, Marenduzzo D, Papantonis A* (2021) RNA polymerase II is required for spatial chromatin reorganization following exit from mitosis. Sci Adv 7: eabg8205.
- Sofiadis K, Josipovic N, Nikolic M, Kargapolova Y, Übelmesser N, Varamogianni I, Zirkel A, Papadionysiou I, Loughran G, Keanes J, Michel A, Gusmao EG, Becker C, Altmüller J, Georgomanolis T, Mizi A, Papantonis A* (2021) HMGB1 coordinates SASP-related chromatin folding and RNA homeostasis on the path to senescence. Mol Syst Biol 17:e9760. [Featured on the issue’s cover]
- Kargapolova Y*, Rehimi R, Kayserili H, Brühl J, Zirkel A, Li Y, Yigit G, Hoischen A, Frank S, Russ N, Trautwein J, Laugsch M, Gusmao EG, Josipovic N, Altmüller J, Nürnberg P, Längst G, Kaiser FJ, Watrin E, Brunner H, Rada-Iglesias A, Kurian L, Wollnik B, Bouazoune K*, Papantonis A* (2021) Overarching control of autophagy and DNA damage response by CHD6 revealed by modeling a rare human pathology. Nat Commun 12:3014.
- Casa V, Gines MM, Gusmao EG, Slotman JA, Zirkel A, Josipovic N, Oole E, van Ijcken WFJ, Houtsmuller AB, Papantonis A*, Wendt KS* (2020) Redundant and specific roles of cohesin STAG subunits in chromatin looping and transcription control. Genome Res 30:515-527.
- Weiterer SS, Meier-Soelch J, Georgomanolis T, Mizi A, Beyerlein A, Weiser H, Brant L, Mayr-Buro C, Jurida L, Beuerlein K, Müller H, Weber A, Tenekeci U, Dittrich-Breiholz O, Bartkuhn M, Nist A, Stiewe T, van IJcken WF, Riedlinger T, Schmitz ML, Papantonis A*, Kracht M* (2020) Distinct IL-1α-responsive enhancers promote acute and coordinated changes in chromatin topology in a hierarchical manner. EMBO J 39:e101533. [Featured in the German Press]
- Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmüller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nürnberg P, Marenduzzo D, Rippe K, Papantonis A* (2018) HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell 70: 730-744. [Featured in F1000]
- Brant L, Georgomanolis T, Nikolic M, Brackley CA, Kolovos P, van Ijcken W, Grosveld FG, Marenduzzo D, Papantonis A* (2016) Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Mol Syst Biol 12: 891. [Highlight of the issue and featured in Nature Methods]
- Frank S, Ahuja G, Bartsch D, Russ N, Yao W, Kuo JC, Derks JP, Akhade VS, Kargapolova Y, Georgomanolis T, Messling JE, Gramm M, Brant L, Rehimi R, Vargas NE, Kuroczik A, Yang TP, Sahito RGA, Franzen J, Hescheler J, Sachinidis A, Peifer M, Rada-Iglesias A, Kanduri M, Costa IG, Kanduri C, Papantonis A, Kurian L (2019) yylncT defines a class of divergently transcribed lncRNAs and safeguards the T-mediated mesodermal commitment of human PSCs. Cell Stem Cell 24: 318-327
- Rada-Iglesias A*, Grosveld FG, Papantonis A*. Forces driving the three-dimensional folding of eukaryotic genomes. Mol Syst Biol 14: e8214 (*co-corresponding authors)
- Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmüller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nürnberg P, Marenduzzo D, Rippe K, Papantonis A (2018) HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell 70: 730-744
- Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmüller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nürnberg P, Marenduzzo D, Rippe K, Papantonis A (2018) HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell 70: 730-744
- Michieletto D, Chiang M, Colì D, Papantonis A, Orlandini E, Cook PR, Marenduzzo D (2018) Shaping epigenetic memory via genomic bookmarking. Nucleic Acids Res 46: 83-93
- Franzen J, Zirkel A, Blake J, Rath B, Benes V, Papantonis A, Wagner W (2017) Senescence-associated DNA methylation is stochastically acquired in subpopulations of mesenchymal stem cells. Aging Cell 16: 183-91
- Brant L, Georgomanolis T, Nikolic M, Brackley CA, Kolovos P, van Ijcken W, Grosveld FG, Marenduzzo D, Papantonis A* (2016) Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Mol Syst Biol 12: 891 (*corresponding author)
- Nikolic M, Papantonis A, Rada-Iglesias A (2016) GARLiC: A bioinformatic toolkit for etiologically connecting diseases and cell type-specific regulatory maps. Hum Mol Genet doi: 10.1093/hmg/ddw423
- Kolovos P*, Georgomanolis T, Koeferle A, Larkin JD, Brant L, Nikolic M, Gusmao EG, Zirkel A, Knoch TA, van Ijcken W, Cook PR, Costa IG, Grosveld FG, Papantonis A* (2016) Binding of nuclear factor kappaB to noncanonical consensus sites reveals its bimodal role during the early proinflammatory response. Genome Res 26: 1478-89 (*corresponding author)
- Melnik S, Caudron-Herger M, Brant L, Rippe K, Cook PR, Papantonis A* (2015) Isolation of the protein and RNA content of the active site of transcription in mammalian cells. Nat Protocols 11: 553-65 (*corresponding author)
- Caudron-Herger M, Cook PR, Rippe K, Papantonis A* (2015) Dissecting the nascent human transcriptome by analyzing the RNA content of transcription factories. Nucleic Acids Res 43: e95 (*corresponding author)
- Kelly S, Georgomanolis T, Zirkel A, Diermeier S, O’Reilly D, Murphy S, Laengst G, Cook PR, Papantonis A* (2015) Splicing of many human genes involves sites embedded within introns. Nucleic Acids Res 43: 4721-32 (*corresponding author)
- Kelly S, Greenman C, Cook PR, Papantonis A* (2015) Exon skipping is correlated with exon circularization. J Mol Biol 427: 2414-7 (*corresponding author)
- Diermeier S, Kolovos P, Heizinger L, Schwartz U, Georgomanolis T, Zirkel A, Wedemann G, Grosveld F, Knock TA, Merckl R, Cook PR, Laengst G*, Papantonis A* (2014) TNFα signaling primes chromatin for NF-κB binding and induces rapid and widespread nucleosome repositioning. Genome Biol 15: 536 (*co-corresponding authors)
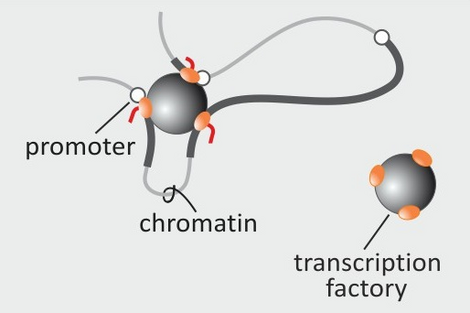
- Papantonis A, Cook PR (2013) Transcription factories: genome organization and gene regulation. Chem Rev doi:10.1021/ cr300513p
- Larkin JD, Cook PR, Papantonis A* (2012) Dynamic configurations of long human genes during a transcription cycle. Mol Cell Biol 32: 2738-2747 (*corresponding author)
For a complete list of publications by our group, please visit: https://www.ncbi.nlm.nih.gov/pubmed/?term=papantonis+a
Lab members

Argyris Papantonis (b. 1978, Athens, Greece) studied Biology at the National and Kapodistrian University of Athens, where he also undertook his PhD studies under the supervision of Prof. Rena Lecanidou. In 2008 he moved to Oxford for his post-doctoral work with Prof. Peter Cook at the Sir William Dunn School of Pathology. In 2013 he was appointed as a Junior Research Group Leader for Chromatin Systems Biology at the CMMC, and in 2018 he was recruited to the Medical Faculty of the Georg-August University of Göttingen where he is currently a W2-Professor for Translational Epigenetics. When he is not doing research he reads, writes, or translates literary texts.
E-mail: argyris.papantonis [at] med.uni-goettingen.de / Twitter: @apapant
Current lab members:
Dr. Athanasia Mizi (Senior postdoc) studied Biology at the National and Kapodistrian University of Athens, where she also undertook her PhD studies under the supervision of Prof. George Rodakis on the molecular evolution of mtDNA. Before joining our lab, Athanasia did her first post-doctoral work with Dr. Paul Brinkkoetter on the role of the key mitochondrial factor, TFAM, in kidney function. Currently, Athanasia is working on senescence-centered 3D chromatin networks; she is funded by the UMG.
E-mail: athanasia.mizi [at] med.uni-goettingen.de
Jinxin Yuan (PhD candidate) studied Clinical Medicine at the Chingqing Medical University, China, and undertook her MSc studies at the Dalian Medical University, China. Before joining our lab, Jinxin worked on the molecular mechanisms regulating H3K4 methylation in hepatoma. Currently, she focuses on the functional dissection of pancreatic cancer proliferation via anti-senescence programs. She is funded by a full PhD fellowship from the China Scholarship Council.
E-mail: jinxin.yuan [at] med.uni-goettingen.de
Dr. Mariano Barbieri (postdoc) graduated in Theoretical Physics from "Federico II" University of Napoli, Italy, where he also did his PhD in statistical models of chromatin architecture with Prof. Mario Nicodemi. Later he spent time as a postdoc in Prof. Ana Pombo's lab in Berlin. He is interested in complex systems behavior and machine learning techniques and joined our laboratory in 2021 after a stint in the private sector (quantitative finance at Morgan Stanley; optics simulations at Italian Space Agency) to develop in silico models of 3D nuclear organization and phase separation. Mariano is supported by the DFG-funded SPP2191 Priority Program on "Molecular Mechanisms of Functional Phase Separation".
E-mail: mariano.barbieri [at] med.uni-goettingen.de
Dr. Adi Mackay-Danieli studied Applied Biology at the University of Applied Sciences of Bonn-Rhein-Sieg, and received her MSc in Molecular Medicine from the University Medical Center Göttingen. Adi did her MSc thesis in our lab as well as her PhD by investigating pancreatic cancer genome regulation in patient-derived organoids. She is associated with the KFO5002 Clinical Research Unit "Deciphering Genome Dynamics for Subtype-Specific Therapy in Pancreatic Cancer".
E-mail: adi.mackay [at] med.uni-goettingen.de
Dr. Anastasios Liakos (postdoc) graduated with a diploma in Biology from the National and Kapodistrian University of Greece, before obtaining an MSc degree in Medical Genetics from the University of Glasgow, UK, and a PhD after working in the laboratory of Dr. Maria Fousteri at BSRC 'Alexander Fleming' in Greece. He is working on the interphase of aging, DNA maintenance, and 3D genome architecture. Anastasios is currently supported by a Human Frontier Science Program postdoctoral fellowship.
E-mail: anastasios.liakos [at] med.uni-goettingen.de
Dr. Andrés Penagos-Puig (postdoc) graduated with a degree in Biology and a Master's in Biochemistry from the National Autonomous University of Mexico, where he also did his PhD in 3D chromatin architecture with Prof. Furlan-Margaril. He is interested in the interplay between RNA polymerases and cohesin and how they shape the genome. Andrés is currently supported by the DFG-funded SFB1565 Collborative Research Center on "Molecular mechanisms and interplay of gene expression processes".
Email: andres.penagos [at] med.uni-goettingen.de
Vassiliki Varamogianni-Mamatsi (PhD candiate) holds a BSc in Biology and a MSc in Molecular Biology and Biomedicine from the University of Crete. Her PhD work focuses on understanding higher-order 3D chromatin architecture using genomics and chromatin simulations. Ilia is funded via the SFB1565 "Molecular Mechanisms and Interplay of Gene Expression Processes" consortium.
E-mail: vassiliki.varamogiannimamatsi [at] med.uni-goettingen.de
Chong Xie (PhD candidate) studied Biotechnology in Hunan University, China, where he also obtained his MSc for Biomedical Engineering. He worked as a research assistant with Dr. Haoyue Zhang at the Shenzhen Bay Laboratory before joining our lab in 2023. His PhD will focus on the connection between 3D genome architecture and phase separation propensity of the transcriptional machinery. Chong is supported by a full PhD fellowship awarded by the China Scholarship Council.
E-mail: chong.xie [at] med.uni-goettingen.de
Markos Tsitsianopoulos (PhD Candidate) studied Biochemistry and Biotechnology at the University of Thessaly, Greece and holds an MSc in Stem Cells and Regenerative Medicine from the Aristotle University of Thessaloniki, Greece. Before joining our lab, Markos worked on the spatial organization of gene expression in breast cancer at the lab of Dr. Christoforos Nikolaou at BSRC 'Alexander Fleming'. His PhD work focuses on dissecting 3D genome architecture in FFPE pancreatic cancer samples. Markos is funded by the KFO5002 Clinical Research Unit.
E-mail: markos.tsitsianopoulos [at] med.uni-goettingen.de
Aaliya Khan (PhD candidate) studied Zoology from Aligarh Muslim University, India and completed her MSc degree in Genetics, Genomics, Epigenetics, and Evolution at Université Paris-Saclay, France. Prior to joining our lab, she worked at the Almouzni lab in Institut Curie, Paris,on the roles of H3.3S31ph in cell cycle progression. Her current work investigates the epigenetic landscape of pancreatic adenocarcinoma cells in response to mechanical cues as part of SCIE-PANC consortium, funded by the TRANSCAN-3 EU research initiative.
E-mail: aaliya.khan [at] med.uni-goettingen.de
Pengfei Yin (PhD candidate) holds a BSc in Aquaculture from the Ocean University of China and an MSc in Systems Biology from the Southern University of Science and Technology, where he began using computational and theoretical methods to study 3D genomics. His PhD research focuses on utilizing computational 3D genomics to better understand the evolution of genomes. He is supported by the DFG-funded RTG2984 EvoREST Program.
E-mail: pengfei.yin [at] med.uni-goettingen.de
Tuba Sena Oğurlu (PhD candidate) received her BSc degree in Molecular Biology and Genetics from Boğaziçi University, Turkey. During this time, she focused on genome-wide analysis of DNA repair at Sabancı University. She later completed her MSc at Bilkent University, Turkey, where she investigated the mechanisms of senescence. Currently, She combines her experience in senescence and DNA repair/damage balance to study the fidelity of gene expression in the presence of DNA/RNA lesions as cells age. Her research is supported by the DFG CRC1678 consortium.
Email: tuba.ogurlu [at] med.uni-goettingen.de
Alumni:
- Dr. Anne Zirkel (former postdoc), currently holding a managerial position in the private healthcare sector.
- Dr. Lilija Brant (former PhD student), currently holding a consultant in the private sector.
- Dr. Milos Nikolic (former PhD student), currently a postdoc with the Peifer lab in Cologne
- Dr. Konstantinos Sofiadis (former PhD student), currently a postdoc with the de Laat lab in Utrecht.
Contact
How to find us? We are currently located on the 2nd floor of the main UMG building (Room 2.E1.216).
Postal address:
AG Papantonis: Translational Epigenetics
Institute of Pathology, University Medical Center,
Georg-August University of Göttingen,
Robert-Koch-Str. 40, 37075 Göttingen, Germany
Tel.: +49 551 39 65734

contact information
- telephone: +49 551 3965734
- e-mail address: argyris.papantonis(at)med.uni-goettingen.de
Das könnte Sie auch interessieren
